The data to be calculated are specified by concatenating the corresponding keywords in the output variable:
- 'ALL': All types of data are written to the corresponding files.
- 'PSA': The atomic and residue solvent accessibilities are
written to the .sol and .psa files, respectively. The algorithm
for the solvent contact areas is described in [Richmond & Richards, 1978]. The
normalization for the fractional areas is carried out as described in
[Hubbard & Blundell, 1987], with the normalization factors courtesy of Simon Hubbard
(personal communication). The single reference is
[Šali & Overington, 1994]. Accessibilities are calculated with scaled radii from
the $MODELS_LIB library, as specified by topology.submodel. The
radii are scaled by energy_data.radii_factor, which should usually be
set to 1.
- 'CAV': The protein and internal cavity volumes are written out.
The calculation on a grid is used. The grid unit is specified by
grid_unit in angstroms (say 1.4). The radii are scaled
by energy_data.radii_factor, which should usually be set to 1. The
cross-sections are written to file file.cav when output contains
CROSS-SECTIONS.
The number_of_steps is the number of small shifts
along x, y, and z that are used in the averaging of the protein and
cavity volumes with respect to small changes in the relative position
of the protein and the grid; the total number of calculations is therefore
equal to the third power of number_of_steps. If orient is True,
the structure is oriented before the volume calculation such that the
moment of inertia are parallel to the x, y, and z coordinate axes (this
orientation minimizes the size of the grid). However, the coordinates of
the MODEL are not changed upon exit from this routine (you need to use
model.orient() to change the orientation of the MODEL).
- 'NGH': Residue neighbors of each residue are listed to a .ngh
file. The MODELLER definition of a residue-residue contact used in
restraints derivation is applied [Šali & Blundell, 1993]: Any pair of residues
that has any pair of atoms within 6Å of each other are in contact.
- 'DIH': All the dihedral angle types defined in the $RESDIH_LIB library (virtual
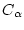 , mainchain, and sidechain
dihedral angles) are written to a .dih file.
, mainchain, and sidechain
dihedral angles) are written to a .dih file.
- 'SSM': Secondary structure assignments are written to a .ssm
file. The algorithm for secondary structure assignment depends on the
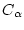 positions only and is based on the distance matrix idea
described in [Richards & Kundrot, 1988]. For each secondary structure type, a `library'
positions only and is based on the distance matrix idea
described in [Richards & Kundrot, 1988]. For each secondary structure type, a `library'
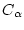 distance matrix was calculated by averaging distance matrices for
several secondary structure segments from a few high resolution protein
structures. Program DSSP was used to assign these secondary structure
segments [Kabsch & Sander, 1983]. Outlier distances were omitted from the averaging. Currently, there
are only two matrices: one for the
distance matrix was calculated by averaging distance matrices for
several secondary structure segments from a few high resolution protein
structures. Program DSSP was used to assign these secondary structure
segments [Kabsch & Sander, 1983]. Outlier distances were omitted from the averaging. Currently, there
are only two matrices: one for the 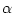 -helix (secondary structure
type 2) and one for the
-helix (secondary structure
type 2) and one for the 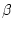 -strand (type 1). The algorithm for
secondary structure assignment is as follows:
-strand (type 1). The algorithm for
secondary structure assignment is as follows:
- For each secondary structure type (begin with a helix, which can
thus overwrite parts of strand if they overlap):
- Define the degree of the current secondary structure fit for each
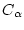 atom by DRMS deviation (
atom by DRMS deviation (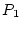 ) and maximal distance
difference
(
) and maximal distance
difference
(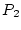 ) obtained by comparing the library distance matrix with
the distance matrix for a segment starting at the given
) obtained by comparing the library distance matrix with
the distance matrix for a segment starting at the given 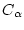 position;
position;
- Assign the current secondary structure type to all
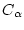 's
in all segments whose DRMS deviation and maximal distance
difference are
less than some cutoffs (
's
in all segments whose DRMS deviation and maximal distance
difference are
less than some cutoffs (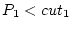 ,
, 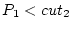 ) and are not
already assigned to `earlier' secondary structure types;
) and are not
already assigned to `earlier' secondary structure types;
- Define the degree of the current secondary structure fit for each
- Split kinked contiguous segments of the same type into separate segments:
Kinking residues have both DRMS and maximal distance difference beyond their respective cutoffs (
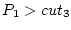 ,
, 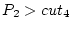 ). The actual single
kink residue separating the two new segments of the same type is the
central kinking residue. Note: we are assuming that there are no multiple
kinks within one contiguous segment of residues of the same secondary
structure type. The kink residue type is
). The actual single
kink residue separating the two new segments of the same type is the
central kinking residue. Note: we are assuming that there are no multiple
kinks within one contiguous segment of residues of the same secondary
structure type. The kink residue type is 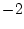 .
.
- If the current secondary structure type is
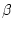 -strand: Eliminate
those runs of strand residues that are not close enough to other
strand residues separated by at least two other residues:
-strand: Eliminate
those runs of strand residues that are not close enough to other
strand residues separated by at least two other residues: 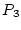 is
minimal distance to a non-neighboring residue of the strand type
(
is
minimal distance to a non-neighboring residue of the strand type
(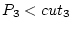 ). Currently, only one pass of this elimination is done,
but could be repeated until self-consistency.
). Currently, only one pass of this elimination is done,
but could be repeated until self-consistency.
- Eliminate those segments that are shorter than the cutoff (
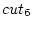 )
length (e.g., 5 or 6).
)
length (e.g., 5 or 6).
- Remove the isolated kinking residues (those that occur on their own or begin or end a segment).
- For each secondary structure type (begin with a helix, which can
thus overwrite parts of strand if they overlap):
- 'CRV': Local mainchain curvatures are written to a .crv
file. Local mainchain curvature at residue
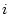 is defined as
the angle between the least-squares lines through
is defined as
the angle between the least-squares lines through 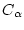 atoms
atoms
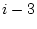 to
to 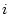 and
and 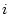 to
to 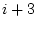 .
.